Identificação de Sistema Eletromecânico - MetaMSS¶
Exemplo criado por Wilson Rocha Lacerda Junior
Procurando mais detalhes sobre modelos NARMAX? Para informações completas sobre modelos, métodos e uma ampla variedade de exemplos e benchmarks implementados no SysIdentPy, confira nosso livro: Nonlinear System Identification and Forecasting: Theory and Practice With SysIdentPy
Este livro fornece orientações detalhadas para apoiar seu trabalho com o SysIdentPy.
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sysidentpy.model_structure_selection import MetaMSS, FROLS
from sysidentpy.metrics import root_relative_squared_error
from sysidentpy.basis_function import Polynomial
from sysidentpy.parameter_estimation import RecursiveLeastSquares
from sysidentpy.utils.display_results import results
from sysidentpy.utils.plotting import plot_residues_correlation, plot_results
from sysidentpy.residues.residues_correlation import (
compute_residues_autocorrelation,
compute_cross_correlation,
)
df1 = pd.read_csv("./datasets/x_cc.csv")
df2 = pd.read_csv("./datasets/y_cc.csv")
df2[5000:80000].plot(figsize=(10, 4))
<Axes: >
(1000, 1)
Decimaremos os dados usando d=500 neste exemplo. Além disso, separamos os dados do MetaMSS para usar a mesma quantidade de amostras na validação de predição. Como o MetaMSS precisa de dados de treino e teste para otimizar os parâmetros do modelo, neste caso, usaremos 400 amostras para treinar em vez de 500 amostras usadas para os outros modelos.
# decimaremos os dados usando d=500 neste exemplo
x_train, x_test = np.split(df1.iloc[::500].values, 2)
y_train, y_test = np.split(df2.iloc[::500].values, 2)
basis_function = Polynomial(degree=2)
estimator = RecursiveLeastSquares()
model = MetaMSS(
xlag=5,
ylag=5,
estimator=estimator,
maxiter=5,
n_agents=15,
basis_function=basis_function,
random_state=42,
)
model.fit(X=x_train, y=y_train)
<sysidentpy.model_structure_selection.meta_model_structure_selection.MetaMSS at 0x229e13e3150>
yhat = model.predict(X=x_test, y=y_test, steps_ahead=None)
rrse = root_relative_squared_error(y_test[model.max_lag :, :], yhat[model.max_lag :, :])
print(rrse)
r = pd.DataFrame(
results(
model.final_model,
model.theta,
model.err,
model.n_terms,
err_precision=8,
dtype="sci",
),
columns=["Regressores", "Parâmetros", "ERR"],
)
print(r)
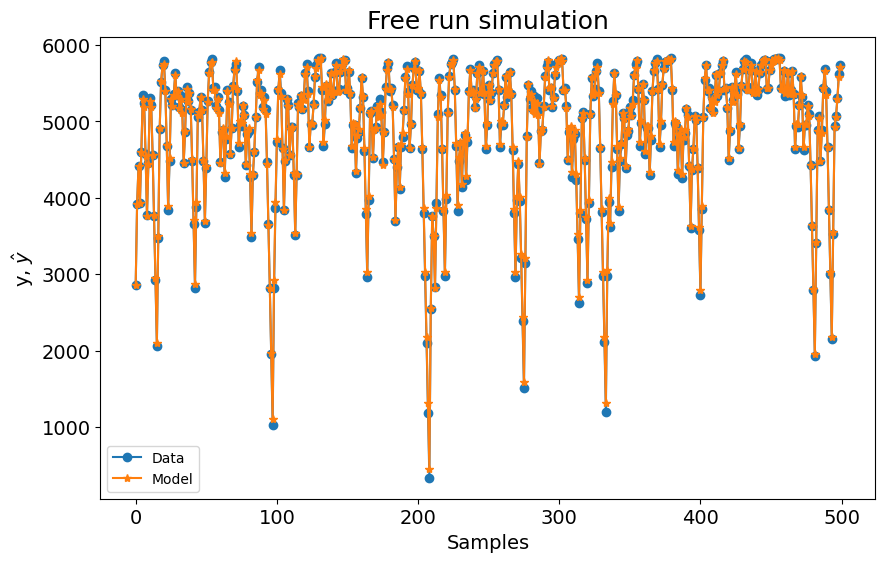
plot_results(y=y_test, yhat=yhat, n=1000)
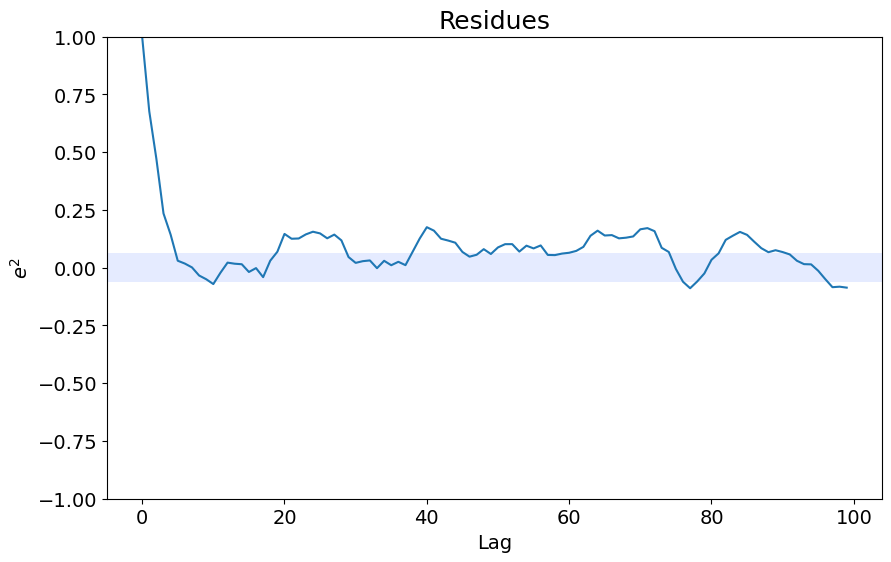
ee = compute_residues_autocorrelation(y_test, yhat)
plot_residues_correlation(data=ee, title="Resíduos", ylabel="$e^2$")
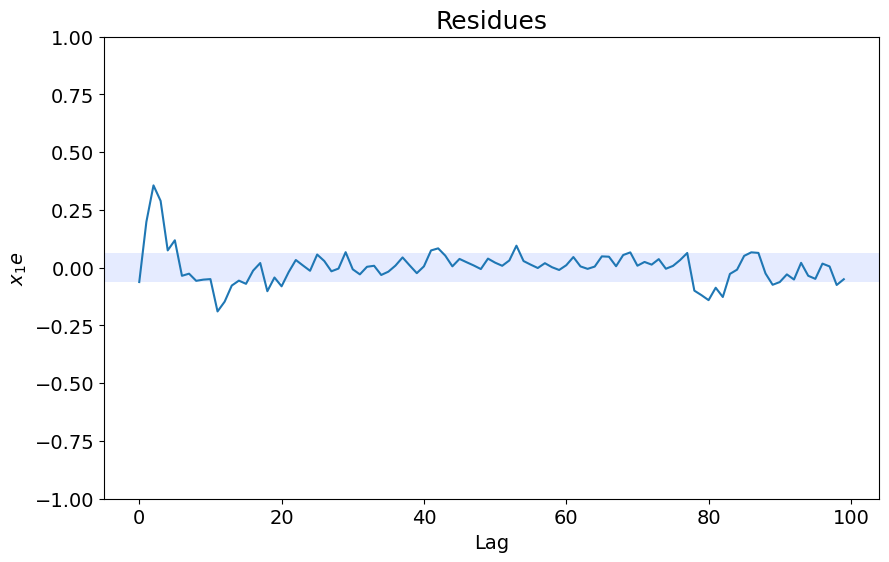
x1e = compute_cross_correlation(y_test, yhat, x_test)
plot_residues_correlation(data=x1e, title="Resíduos", ylabel="$x_1e$")
0.035919583498004094
Regressores Parâmetros ERR
0 1 -6.1606E+02 0.00000000E+00
1 y(k-1) 1.3117E+00 0.00000000E+00
2 y(k-2) -3.0579E-01 0.00000000E+00
3 x1(k-1) 5.7920E+02 0.00000000E+00
4 x1(k-3) -1.8750E-01 0.00000000E+00
5 x1(k-1)y(k-1) -1.7305E-01 0.00000000E+00
6 x1(k-2)y(k-1) -1.1660E-01 0.00000000E+00
7 x1(k-1)y(k-2) 1.2182E-01 0.00000000E+00
8 x1(k-2)y(k-2) 3.4112E-02 0.00000000E+00
9 x1(k-1)y(k-3) -4.8970E-02 0.00000000E+00
10 x1(k-1)y(k-4) 1.3846E-02 0.00000000E+00
11 x1(k-2)^2 1.0290E+02 0.00000000E+00
12 x1(k-3)x1(k-2) 8.6745E-01 0.00000000E+00
13 x1(k-4)x1(k-2) 3.4336E-01 0.00000000E+00
14 x1(k-5)x1(k-2) 2.7815E-01 0.00000000E+00
15 x1(k-3)^2 -9.3749E-01 0.00000000E+00
16 x1(k-4)x1(k-3) 6.1039E-01 0.00000000E+00
17 x1(k-5)x1(k-3) 3.9361E-02 0.00000000E+00
18 x1(k-4)^2 -4.6335E-01 0.00000000E+00
19 x1(k-5)x1(k-4) -9.5668E-02 0.00000000E+00
20 x1(k-5)^2 3.6922E-01 0.00000000E+00
0.0017530517788608157
from sklearn.neighbors import KNeighborsRegressor
from sklearn.svm import SVC
from sklearn.tree import DecisionTreeRegressor
from sklearn.ensemble import RandomForestRegressor
from catboost import CatBoostRegressor
from sklearn.linear_model import ARDRegression
from sysidentpy.general_estimators import NARX
xlag = ylag = 5
estimators = [
(
"NARX_KNeighborsRegressor",
NARX(
base_estimator=KNeighborsRegressor(),
xlag=xlag,
ylag=ylag,
basis_function=basis_function,
),
),
(
"NARX_DecisionTreeRegressor",
NARX(
base_estimator=DecisionTreeRegressor(),
xlag=xlag,
ylag=ylag,
basis_function=basis_function,
),
),
(
"NARX_RandomForestRegressor",
NARX(
base_estimator=RandomForestRegressor(n_estimators=200),
xlag=xlag,
ylag=ylag,
basis_function=basis_function,
),
),
(
"NARX_Catboost",
NARX(
base_estimator=CatBoostRegressor(
iterations=800, learning_rate=0.1, depth=8
),
xlag=xlag,
ylag=ylag,
basis_function=basis_function,
fit_params={"verbose": False},
),
),
(
"NARX_ARD",
NARX(
base_estimator=ARDRegression(),
xlag=xlag,
ylag=ylag,
basis_function=basis_function,
),
),
(
"FROLS-Polynomial_NARX",
FROLS(
order_selection=True,
n_info_values=50,
ylag=ylag,
xlag=xlag,
basis_function=basis_function,
info_criteria="bic",
err_tol=None,
),
),
(
"MetaMSS",
MetaMSS(
norm=-2,
xlag=xlag,
ylag=ylag,
estimator=estimator,
maxiter=5,
n_agents=15,
loss_func="metamss_loss",
basis_function=basis_function,
random_state=42,
),
),
]
all_results = {}
for model_name, modelo in estimators:
all_results["%s" % model_name] = []
modelo.fit(X=x_train, y=y_train)
yhat = modelo.predict(X=x_test, y=y_test)
if model_name in ["FROLS-Polynomial_NARX", "MetaMSS"]:
result = root_relative_squared_error(
y_test[modelo.max_lag :], yhat[modelo.max_lag :]
)
else:
result = root_relative_squared_error(y_test, yhat)
all_results["%s" % model_name].append(result)
print(model_name, "%.3f" % np.mean(result))
NARX_KNeighborsRegressor 1.158
NARX_DecisionTreeRegressor 0.203
NARX_RandomForestRegressor 0.146
NARX_Catboost 0.120
NARX_ARD 0.083
FROLS-Polynomial_NARX 0.057
MetaMSS 0.036
for model_name, metric in sorted(
all_results.items(), key=lambda x: np.mean(x[1]), reverse=False
):
print(model_name, np.mean(metric))
MetaMSS 0.035919583498004094
FROLS-Polynomial_NARX 0.05729765719062527
NARX_ARD 0.08265856190495872
NARX_Catboost 0.12034851661643597
NARX_RandomForestRegressor 0.14557973585496042
NARX_DecisionTreeRegressor 0.203057724881072
NARX_KNeighborsRegressor 1.157787546845798